You may have seen a recent paper by Yuan Hu and Ingo Mügge from Alkermes on ‘In Silico Positional Analogue Scanning with Amber GPU-TI’. The idea behind positional analogue scanning (PAS) is relatively simple: you have a hit molecule, and you’d like to know what changes (and in what directions) are likely to lead to an increase in potency. Would adding something in the 3-position be a good idea? If so, what sort of functional group should we add? Traditionally this relied largely on the skill and judgement of expert chemists to decide which vectors looked good (both from a synthetic feasibility point of view and from any pre-existing knowledge of the SAR), and what changes to make (often following, either directly or in spirit the venerable Topliss Tree).
However, the rise of accurate free energy methods in recent years provides an alternative way to approach the problem. Relative binding free energy calculations allow us to compute with a fair degree of accuracy whether one molecule is likely to bind more strongly than a related one, and the more similar the two molecules are, the more accurate the calculations tend to be. PAS is thus something of a best-case scenario as we have very small and restricted changes being made to the parent molecule, which means that our RBFE predictions are going to be as accurate as they get!
A quick, easy and accurate method to explore SAR with confidence
This concept has been built into the upcoming release of Flare™ V6.1, as a component called Hit Expander. The workflow is that you select a molecule and select a set of allowed modifications to be applied either to the whole molecule or to a selected substructure. Hit Expander will then automatically generate all the resulting analogues and will deduplicate them and sort out charge states and initial conformations for you. You can then take the resulting set of ideas into Flare FEP to get an accurate ranking of which ideas are good, and which not so good.
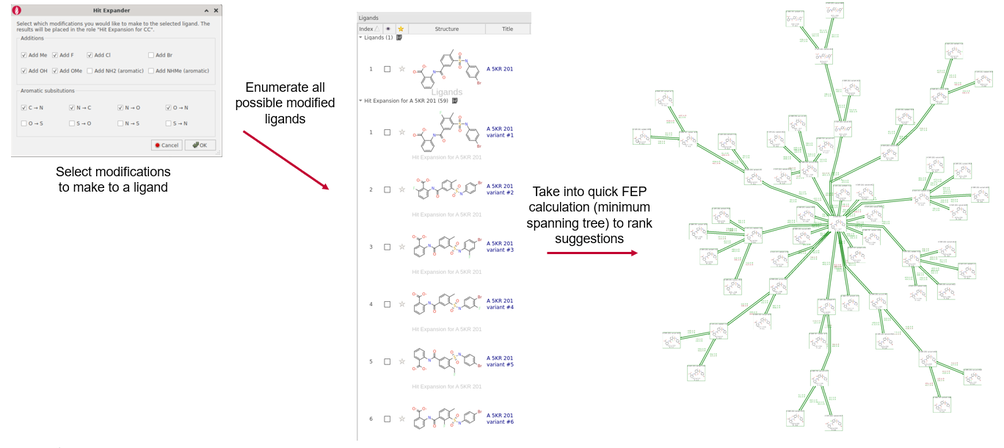
Figure 1. The Hit Expander workflow.
RBFE calculations can be expensive, and the Hit Expander workflow can generate 70 or 80 variations on the average druglike molecule, which would be a significant computing cost for a standard FEP calculation. However, when working on the validation for Hit Expander it became apparent that there were a number of significant time savings that could be made. Firstly, since the transformations are all small there’s limited benefit to trying to apply cycle closure correction. As a result, you can use a minimum spanning tree (as illustrated in Figure 1) rather than a fully-connected network. In addition, although we would normally run each link in both directions, for robustness and to provide error statistics, this is not needed in the Hit Expander workflow.
Moreover, because the transformations are small, the RBFE simulations converge very quickly. Normal Flare FEP runs do 4ns per lambda window, with an average of 10 lambdas per link, so around 40ns of simulation time per link. However, looking at a wide variety of data sets it became apparent that you could drastically reduce this time with only a small loss of accuracy. As can be seen in Figure 2, using 1ns rather than 4ns has only a small impact, and even going to 0.5ns or shorter still provides a good correlation with the 4ns calculations.
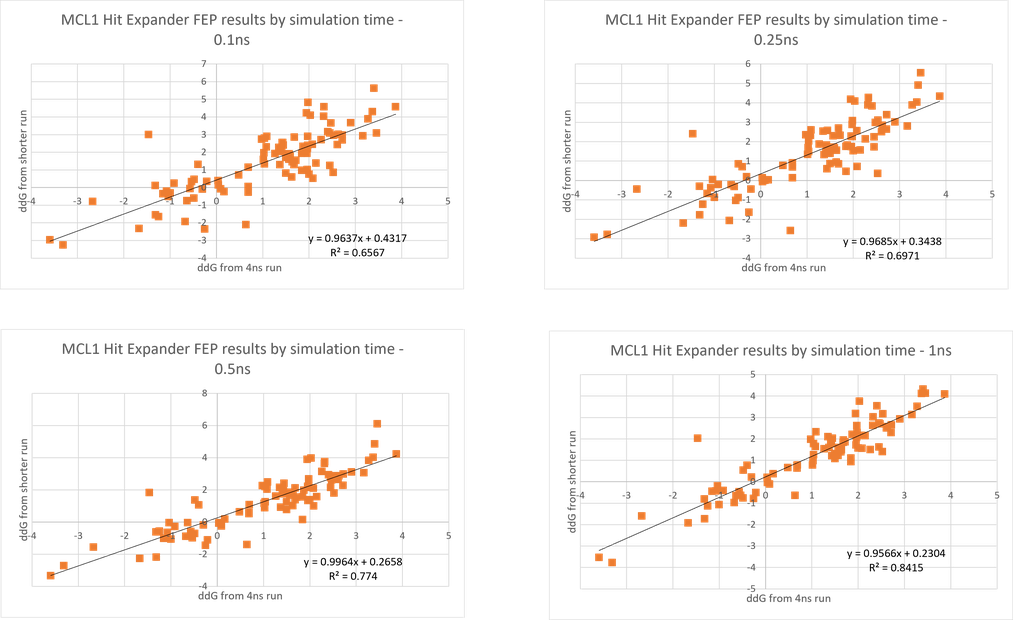
Figure 2. Correlation of ddG values from short runs with the values from 4ns runs.
Taken together, these measures drastically reduce the amount of calculation needed to evaluate the Hit Expander results. For example, the FEP calculation needed to rank the Hit Expander results on a small protein running at 1ns takes only roughly 20 GPU-minutes per molecule (on a mid-range sub-$1000 GPU). This means that you can rank a set of 80 Hit Expander results in only 20-30 GPU-hours in total. The process is also highly parallelisable, with a minimum time-to-answer for a single molecule of around 8 minutes if you have 10 or more GPUs available (including job submission delays and so on).
So, even if you have only very limited GPU resources, Hit Expander will allow you to rank a wide variety of small changes to your lead molecule in less than 24 hours and give excellent guidance not only on what positions are likely to lead to improved activity when substituted, but what types of substitutions are favoured. Furthermore, at only a small extra computational cost you could take the top 5 or so ideas from the quick calculation and put them into a Flare FEP experiment run with the more accurate ‘Normal’ settings, to get additional reassurance as to the degree of increase in activity to be expected from these favoured changes. The results from Hu and Mügge suggest that in all the cases where their workflow predicted an improvement in affinity of a factor of 10 or more, the calculation was correct about the direction (the affinity did actually improve), and our preliminary results are showing the same sort of success rates.
Of course, evaluation of Hit Expander results could be performed in other ways too including regression or classification QSAR models, Electrostatic Complementarity™, or simply visualizing the proposed poses. Indeed, combining ligand-based and structure-based techniques with multiple Hit Expander experiments is an excellent way to rapidly assess and step through the hit-to-lead process with minimal expenditure on wet-chemistry.
Hit Expander coming soon in Flare V6.1
Hit Expander is a new feature in Flare V6.1, due for release this autumn. Subscribe to our newsletter to be amongst the first to hear when it’s released.