Drug discovery research often involves making small
modifications to a compound and studying the subsequent effect on
activity. Once a closely related series of compounds has been made and
tested, it is very important to analyze which of the substituents
enhances activity, and to also know which substitution positions
significantly affect the activity, in order to develop the next
synthetic strategy. R-group decomposition and analysis in Flare™
is a method that allows you to classify substituents by their position
in a given core structure and helps you to identify active substituents
at each position.
If the molecules in the dataset have activity data, you can look at
the distribution of activity values for each substituent at each
substitution position. If you look at two substitution positions at the
same time, you can obtain a matrix of substituents, which provides more
useful information for identifying the substitution pattern found in the
most-active structures in the series, as well as gaps in the chemical
exploration strategy.
In this example, we show how R-group Analysis is performed in Flare
using a set of CDK9 inhibitors collected from ChEMBL. To simplify the
data, the IC50, EC50, AC50, Ki, and Kd (nM) activity values reported by
ChEMBL have been treated as one median activity value for each molecule.
We excluded molecules of MW > 500 or SlogP > 5 that would
interfere with the analysis and prepared a data set of 780 compounds in
total.
Defining the Scaffold for R-group Decomposition
The most active compound in this dataset has a 4-amino-pyrimidine
skeleton (CHEMBL3694408), with pActivity=11.3. This represents a good
starting point to perform an R-group decomposition analysis on.
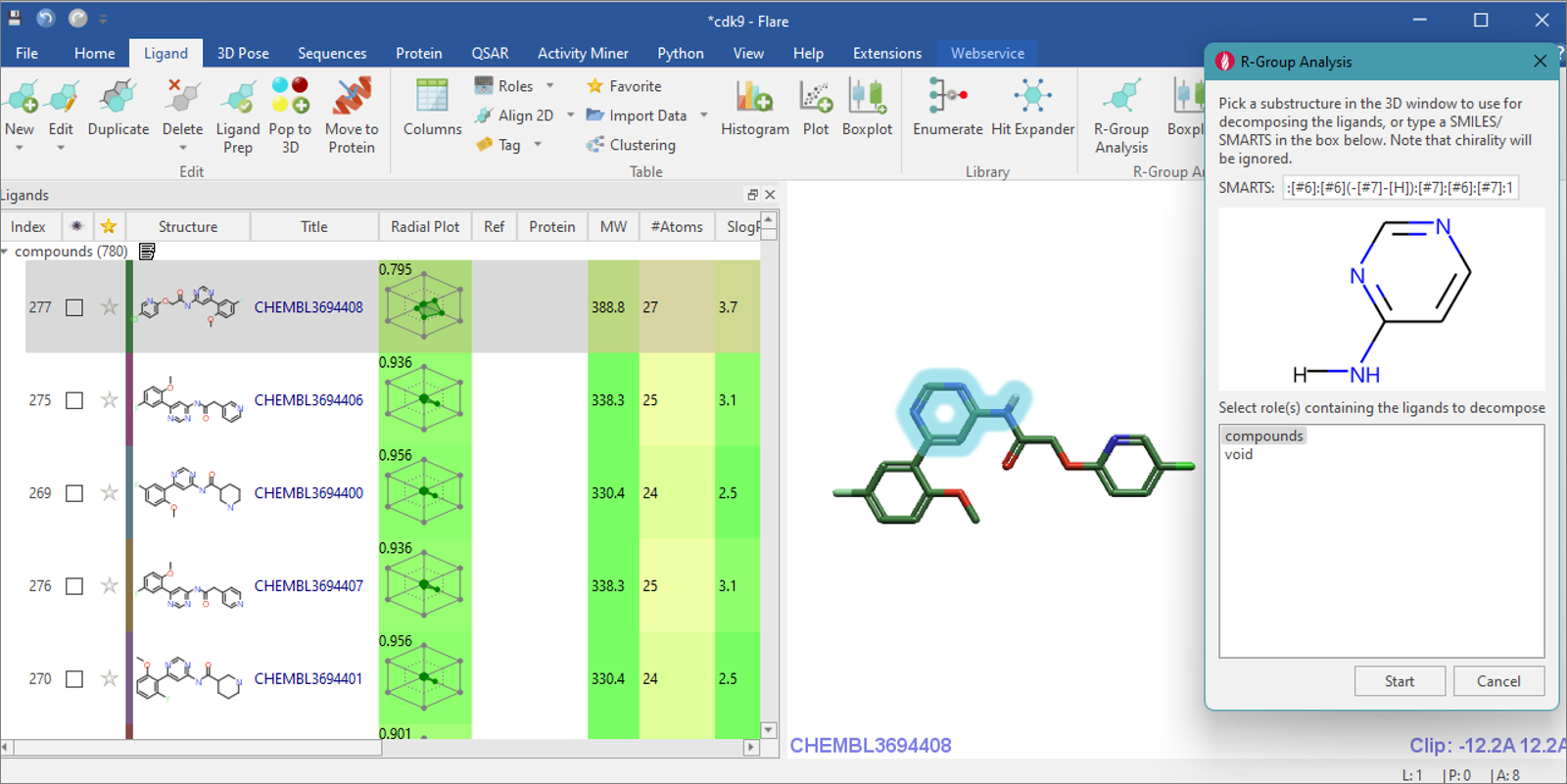
Figure 1. R-group Analysis dialog and selection of atoms in 3D window.
Click on the ‘R-group Analysis’ icon from
the ‘Ligand’ tab, which will open the R-group Analysis panel. In the 3D
window, select the aminopyrimidine moiety and confirm the picked atoms
are depicted in the panel before clicking ‘Start’. Only molecules
matching this substructure in the specified ligand roles will be
included in the subsequent R-group decomposition analysis.
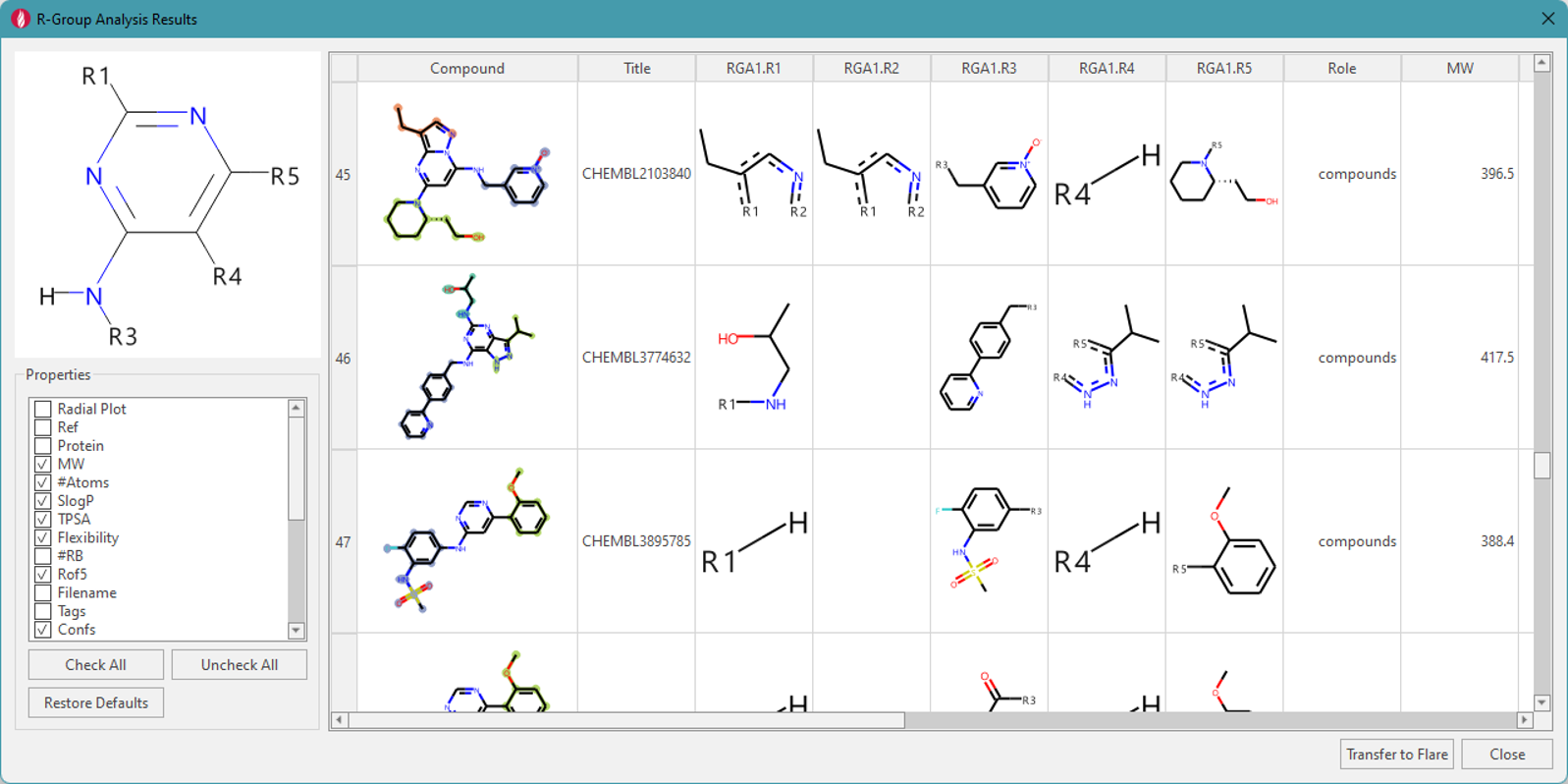
Figure 2. Decomposition dialog: analyzed
substitution positions (left top), property filter (left bottom), and a
table of molecules showing the core with a different color used for each
R-group.
72 compounds with a matching substructure
were found in this dataset. The analysis also indicated that the
attached R-groups in this ‘mini-series’ of molecules can be found at any
of the 4 identified locations: R1, R4, and R5 on the pyrimidine ring,
and R3 on the exocyclic amino group (Figure 2, top left). R2, which is
missing in this pattern, is used internally as a placeholder for
bicyclic substitution, and is shown in the table view. Other columns in
the table view include the ‘Compound’ image, where each decomposed
substituent is colored for specific position, while the checked molecule
properties are shown in subsequent columns. The results of this
decomposition can be transferred to the main ‘Ligands Table’ by clicking
on ‘Transfer to Flare’ button. Columns in the ‘Ligands Table’ starting
with ‘RGA1.’ are the results of this first R-group decomposition
analysis (Figure 3). Subsequent R-group decompositions will have columns
beginning with ‘RGA2.’, ‘RGA3.’ etc.
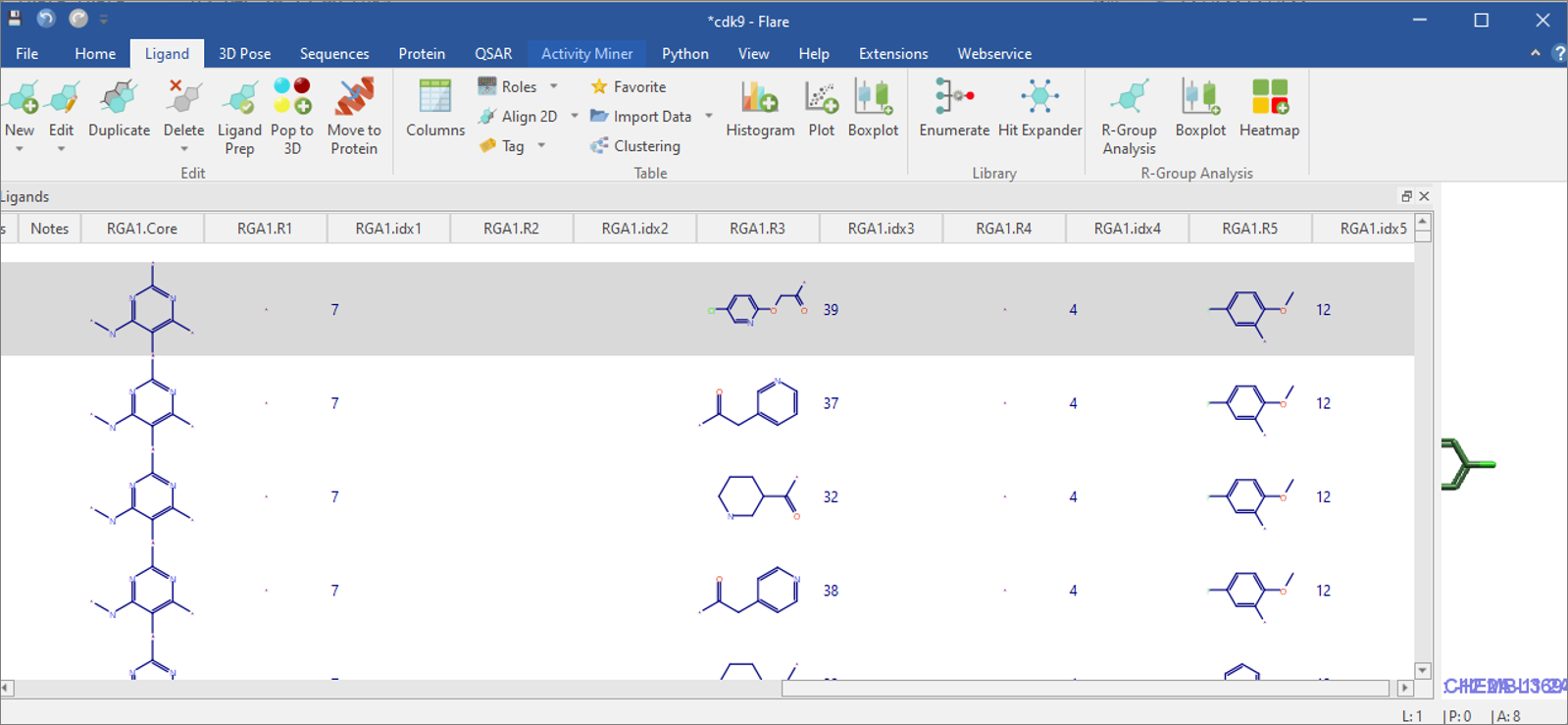
Figure 3. The Flare ‘Ligands Table’ has R-group decomposition results in columns.
Analyzing the R-group Decomposition Results using Boxplots
R-group Analysis boxplots enable you to examine the activity
distribution of the various R-groups at a single substitution position.
Figure 4 shows a boxplot of R5, with the second column from the left
showing that compounds with an unsubstituted Hydrogen have an activity
range of 5.7-8.8 (where the 25th percentile quartile, median, and 75th
percentile quartile are shown within the box). When hovering over each
of the individual points, an image showing the 2D structure of the
molecule with its activity will pop up.
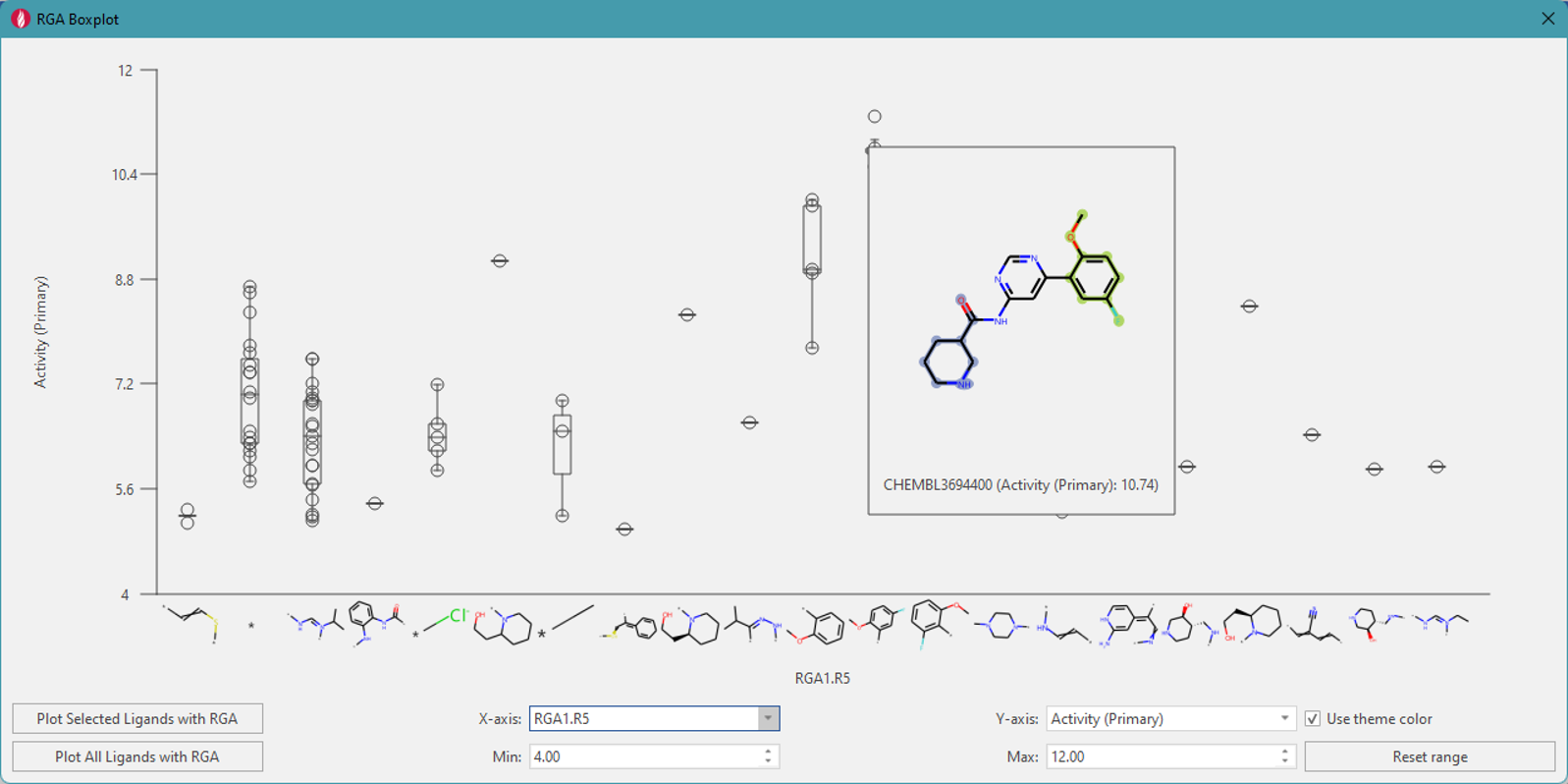
Figure 4. Boxplot for substitution position R5 with activity ranges.
The boxplot shows that the
fluoromethoxyphenyl group substitution at R5 significantly contributes
to the activity. As well as selecting individual points, you can also
specify a rectangular region in the boxplot to select multiple ligands
(Figure 5).
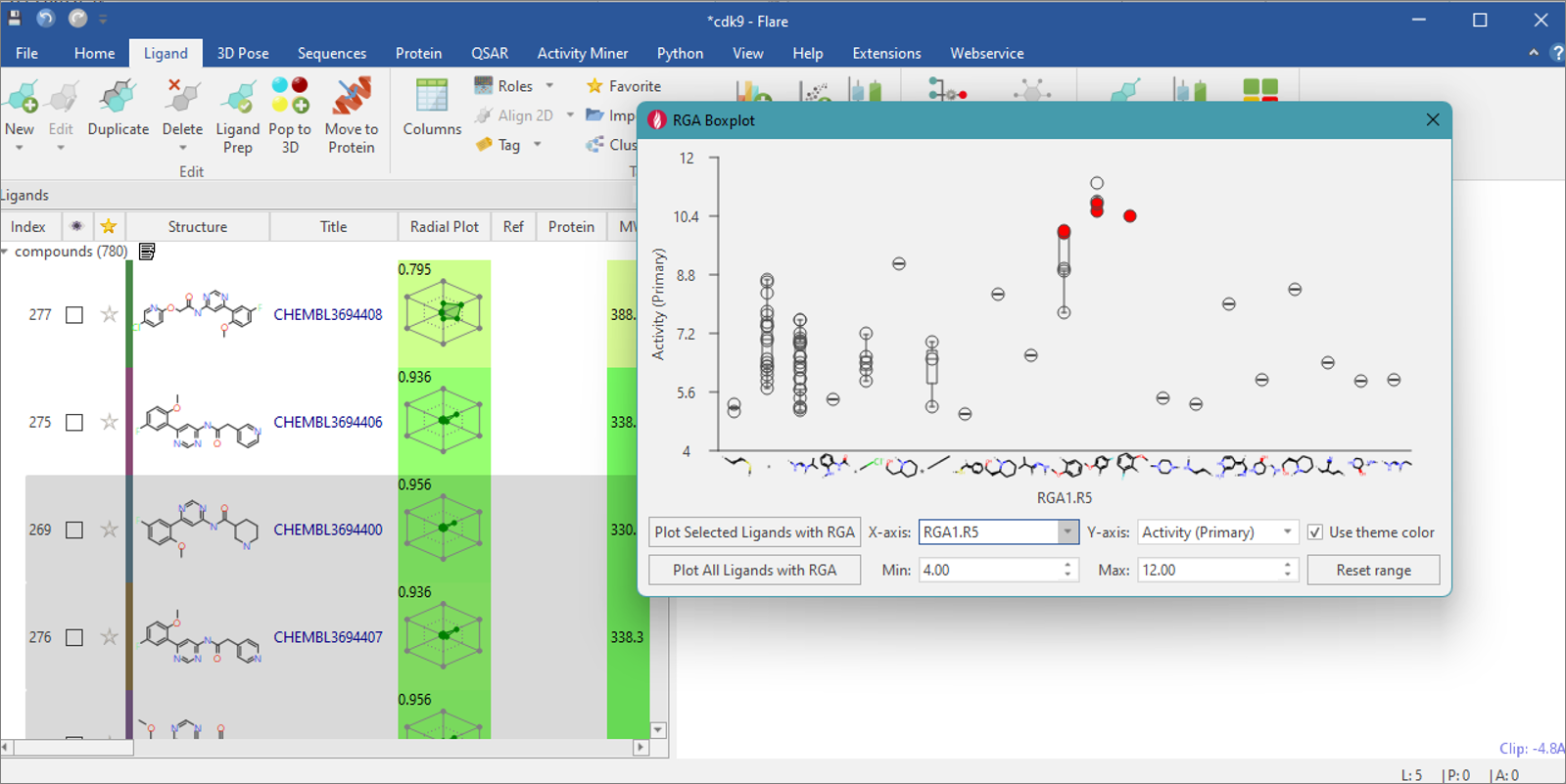
Figure 5. Rectangular region highlights molecules in the main ‘Ligands Table’.
Analyzing the R-group Decomposition Results Using Heatmaps
Another way to review the data is by using a heatmap which is
color-coded by activity. Figure 6 shows an example of a two-dimensional
matrix with R5 (horizontal) and R4 (vertical). The color of the cell
(which corresponds to the color bar at the top of the RGA Heatmap
window) indicates whether the presence of both substituents has a
positive or negative effect on the activity. For example, the
strong-green cell where the mouse pointer is located has four compounds
with a high median activity value. This cell identifies the group of
compounds with high activity shown in the pop-up window; clicking on the
cell selects these compounds in the ‘Ligand Table’ (in Flare’s main
window). The compound shown in the lower right corner is the compound
with the highest activity value in the original dataset, and you can
easily identify that there are three other compounds with the same core
structure and substitution pattern that also have high activity.
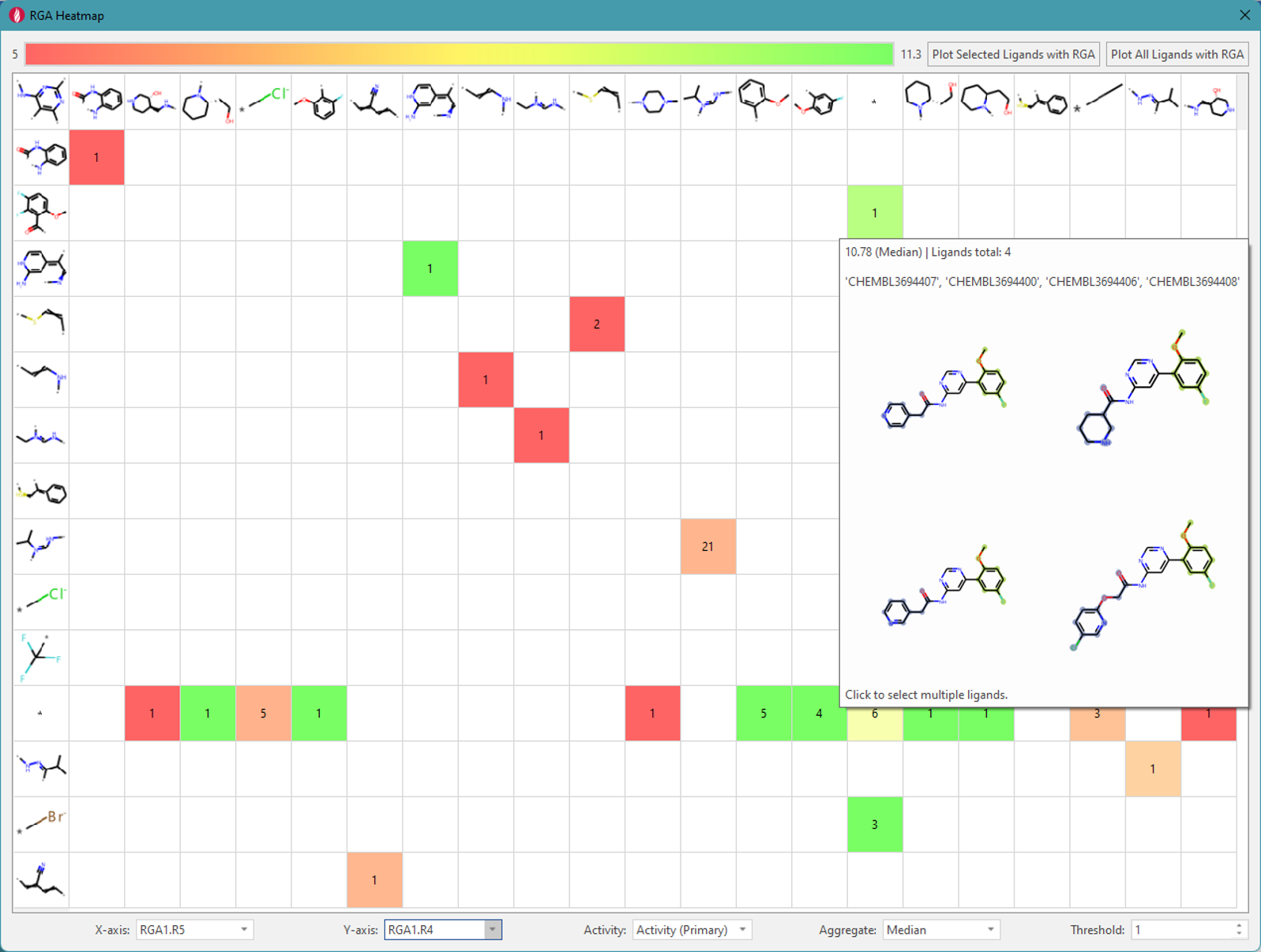
Figure 6. Heat map (matrix) for two specified substitution positions.
Clicking on a substituent structure on the
top row of the matrix will sort the column by its frequency and the
aggregated activity. Figure 7 shows the first part of a matrix after
clicking on three of the R5 substituents to collect the structures
containing them towards the top of the matrix. This gives you an idea of
how sparse the matrix is with any of the substituents present in the
most-active compounds, and helps you subsequently identify further
possible changes to existing compounds which don’t contain these
favorable groups. The check marks in Figure 7 highlight specific groups
that could be added to existing molecules to potentially improve their
activity.
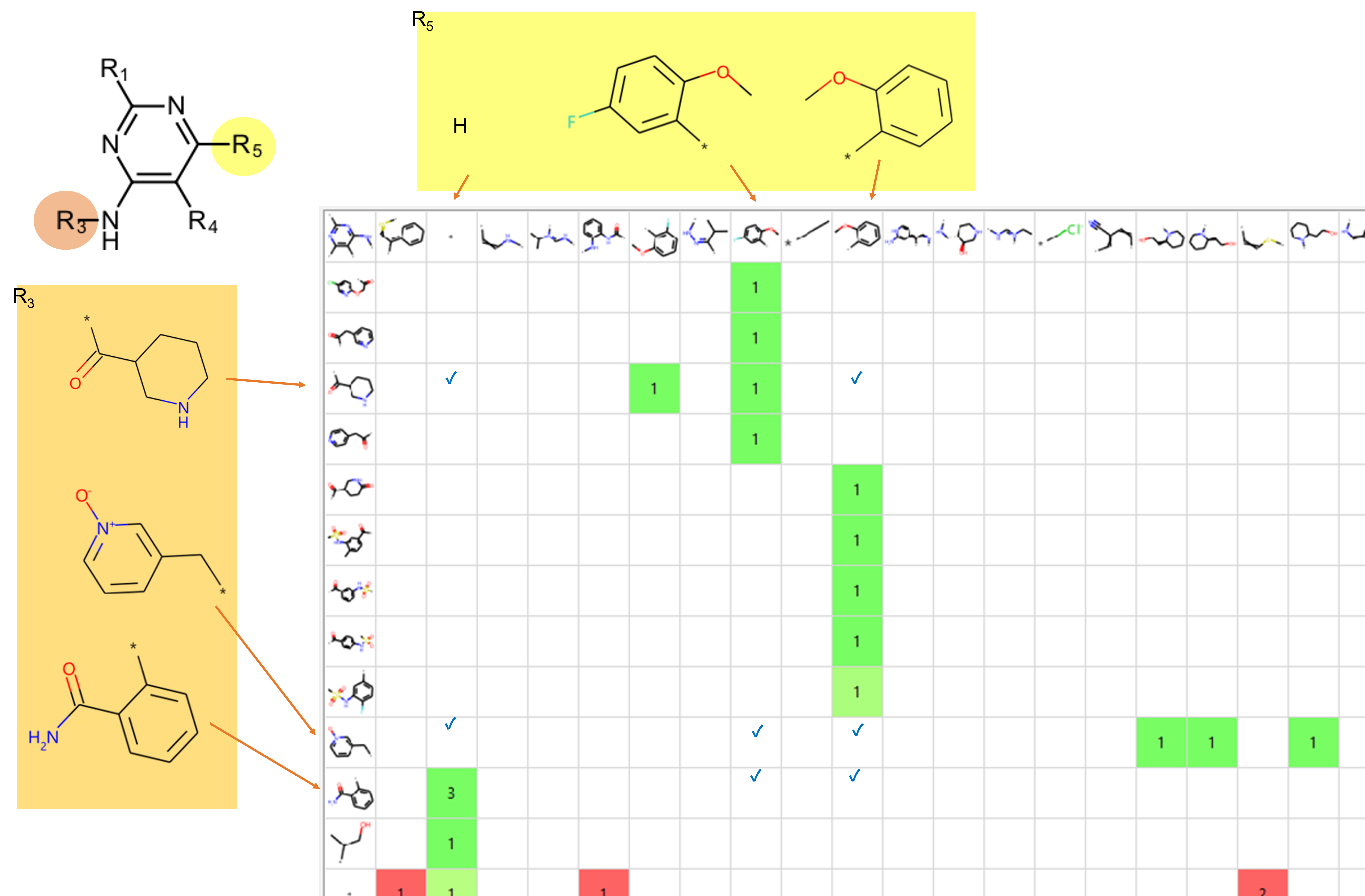
Figure 7. Part of sorted matrix for finding missing compounds.
Conclusion
R-group Analysis enables you to examine the distribution of active
compounds by substituent position, and to understand the structural
features required for high activity, which is useful for designing
active molecules that may have been missed in the initial medicinal
chemistry design and synthesis stage. In the Flare GUI, users can easily
perform R-group analysis using only a few mouse clicks.
Begin to analyze your molecule series with the R-group decomposition functionality, by requesting a free evaluation of Flare today!