The key to designing and executing a successful virtual screen is to use all the available information on the target being considered, and to explore as much diverse chemical space as possible when selecting compounds for purchase. In this study, we used a combination of ligand-based virtual screening with Blaze™ and information about the protein structure from Flare™ to find ligands that could be potentially used as appropriate FOXO-peptide mimetics.
Introduction
The approach taken to targeting biomolecular structures in a drug discovery project is dependent on what binding partners are involved (Figure 1). The most common approach is to target the binding site of a known modulator by introducing a ligand which competes for the same site. However, there are many biomolecules that don’t have obvious small molecule modulators but do interact with other biomolecules such as nucleic acids (DNA and RNA) or proteins. In the latter case, protein-protein interactions usually occur over a significant surface area of the two interacting proteins. The interaction energy between the two molecules is gained from many different smaller contacts over the larger surface area, rather than being concentrated in a small region, such as a binding site. This makes targeting these interactions difficult with a small molecule because there are usually not enough interactions available for small molecules to strongly bind and compete with the larger protein.
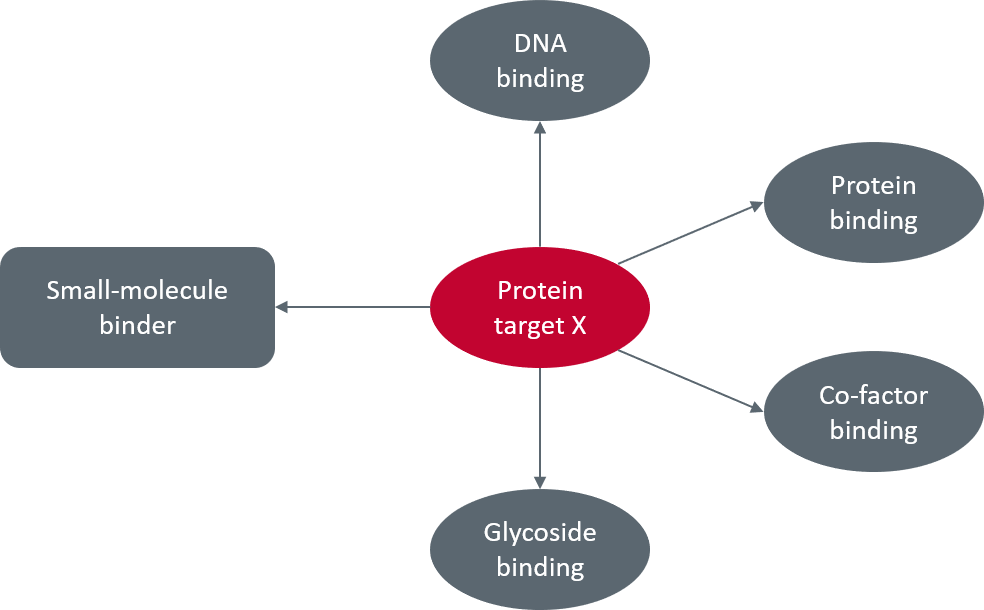
Figure 1. Possible partners for protein target X.
Despite this, there are situations where a small portion of the PPI interface is responsible for the majority of the binding energy between the two partners. Lab-based studies can be done where smaller peptide probes which mimic sections of the protein partner can be used to determine the importance of various regions for binding. If it is possible to identify a smaller region that provides a significant contribution, then it may be possible to target this region with a small molecule.
When the target region has been identified, the next stage is to start looking for possible small molecules which ‘mimic’ the original peptide. One approach is virtual screening, which takes known information about the binding region and looks for molecules which can make favorable interactions with the region of interest. With Cresset’s virtual screening platform, Blaze, the electrostatic, hydrophobic and Van-der-Waals features of the query peptide are used to search for commercially available compounds that have similar features.
Finding new small molecule leads for Forkhead box proteins (FOXO)
FOXOs are transcription factors that have a N-terminal DNA-binding domain. They recognize many different protein partners, such as 14-3-3 proteins. Disruption of FOXO1 pathways can result in several metabolic diseases including diabetes, while FOXO3 has been associated in different tumor entities, promotes tumor angiogenesis, and is involved in apoptosis. Therefore, the FOXO protein family appears to be a good set of biomolecules to target in a drug discovery project. However, their key interactions are with other biomolecules such as DNA, so it is one of the more complex systems to study.
The first stage of this project was to collate the available data associated with the FOXO proteins. No known small molecules were found, but FOXOs are transcription factor proteins and therefore bind with DNA. After searching for the appropriate protein sequence through UniProt, available PDB structures were identified for the FOXO family of proteins.
Targeting the DNA-binding domain
Structures were found which showed the interaction between the FOXO proteins and DNA. In Cresset’s drug discovery platform, Flare, we were able to align and superimpose the available crystal structures of the DNA-binding domain of the FOXO protein family (Figure 2a). After completing the superimposition, it became clear that there were very few specific interactions for a small, ligand-size molecule to utilize when binding to the FOXO, while in addition, any potential binding site is very limited in size (figure 2b). Therefore, an alternative approach needed to be considered.

Figure 3. The 6QZR crystal structure of the 14-3-3 protein (light brown) with a phospho-peptide fragment of FOXO1 (pink).
The aim was to use Blaze to target the FOXO-binding site of the 14-3-3 protein but do this by considering the structural features of the FOXO peptide present in the crystal structure. Taking the central four residues of the FOXO peptide, it was possible to identify the key features of this part of the molecule (Figure 4) – i.e., the regions of positive charge (red spheres), negative charge (blue spheres), the hydrophobic features (orange spheres) and shape-based features (yellow spheres).

Figure 4. The field points which describe the molecular features of the FOXO peptide.
The Blaze interface (which can be accessed through a web-based portal, or through Flare) also allows the scientist to guide the ligand search by introducing pharmacophore or field-point constraints, which is useful if known specific interactions or features need to be present in the identified ligands. When running a virtual screen within Blaze, it is possible to restrict the database of commercially available compounds being searched by attributes such as specific suppliers or by molecular weight. In the Cresset Blaze collection, there are currently around 20 million molecules which can be searched (Figure 5).
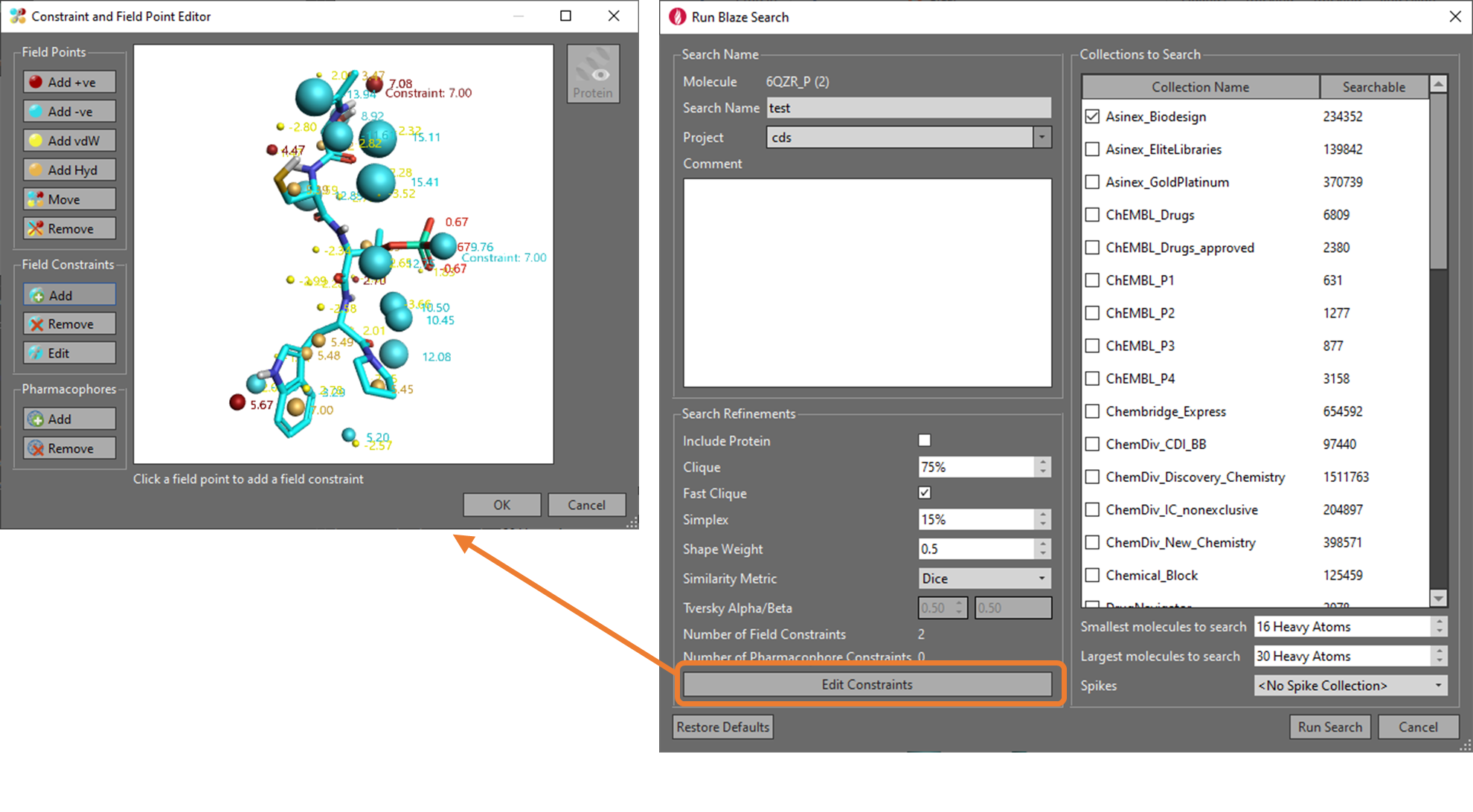
Figure 5. From the Flare interface, it is possible to set-up the input parameters for Blaze. For example, it is possible to guide the virtual screening experiment using field and/or pharmacophore constraints, and define the supplier databases and size of molecules which are going to be searched.
The top-scoring hits were downloaded into Flare and viewed within the 14-3-3 receptor. Although none of the 14-3-3 protein was used to guide the virtual screen, the query was taken from the 6QZR crystal structure thus placing the virtual hits in the same reference frame as the 14-3-3 protein.
Once the top-scoring hits were downloaded into Flare, it was possible to do further post-processing and analysis (Figure 6), such as, visual inspection to eliminate ligands which, for example, look too strained; docking into the 14-3-3 protein; and investigating how well the new ligands complement the binding site electrostatically. In Flare, it is also possible to identify molecules which have ideal ranges in physical properties such as TPSA, logP or flexibility and therefore ensure that only molecules with the appropriate calculated properties are selected. When purchasing compounds from a virtual screening experiment, it is important to ensure that chemical space is sufficiently sampled. In the forthcoming release of Flare V5, it will also be possible to cluster the results to aid compound selection.
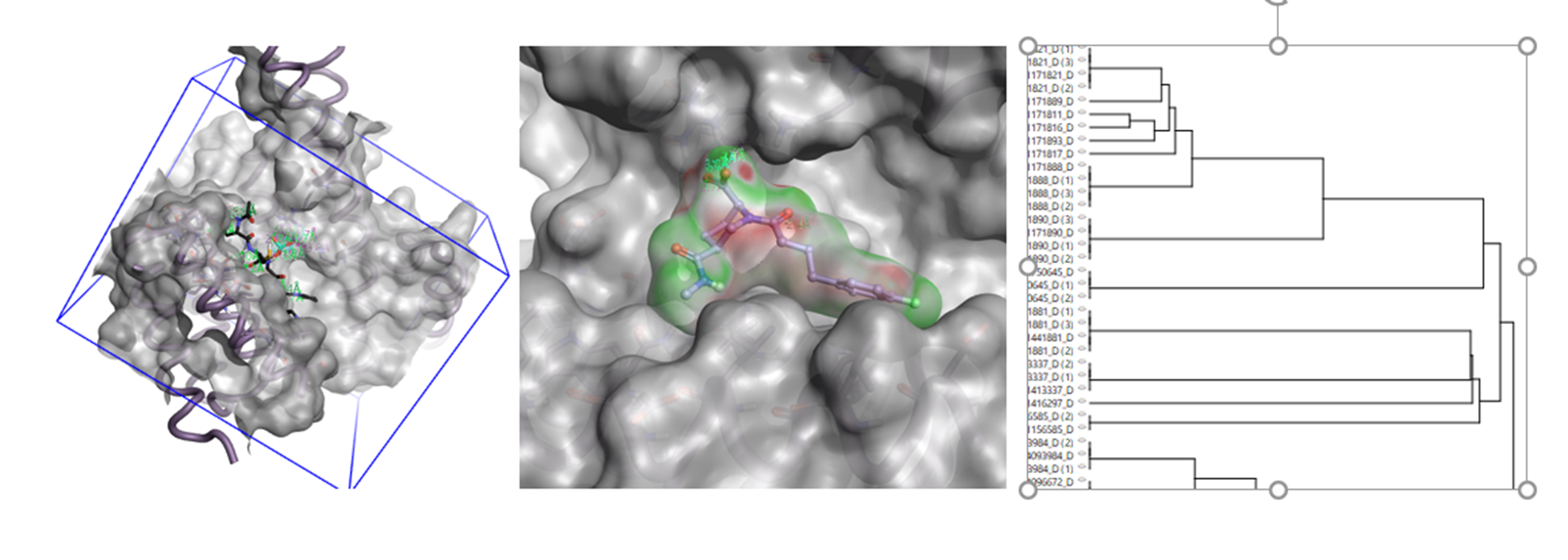
Figure 6. Graphical representations of Flare features which could be used to guide selection of compounds obtained from a Blaze virtual screen. (a). Docking, (b). Electrostatic Complementarity™ and (c). Clustering.
Figure 7 shows some examples of the output from Blaze. The top-left most molecule (1) is the query peptide and many of the shown examples interact with the phosphate recognition site using an acidic group, but some of the molecules don’t (6,9), which would make for an interesting investigation.
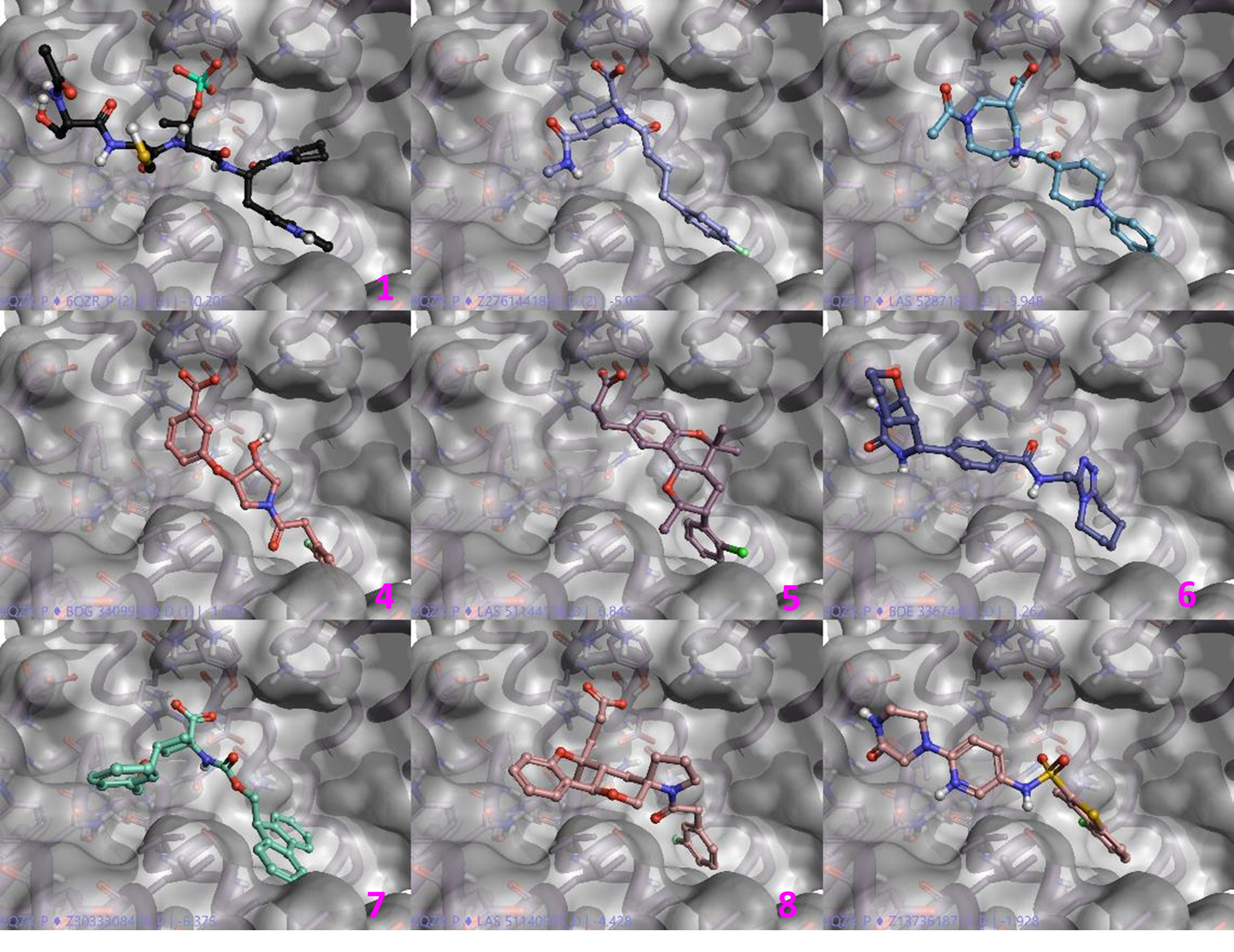
Figure 7. Some of the output from the Blaze virtual screen as shown in Flare.
Conclusion
A combination of ligand-based and structure-based approaches can be used to exploit all the target information available when designing a virtual screening strategy. A well-designed search has the capability of identifying small molecule hits for more-challenging targets that interact with other biomolecules such as nucleic acids (DNA and RNA) or proteins.