Running Molecular Dynamics (MD) calculations with membrane proteins is one of the new features recently introduced within Flare.
Not only do membrane proteins play a key role in a cell’s biology, but
around 20% of the human genome code for them and with regards to
therapeutic targets they are of significant interest.[1] Classes of
membrane proteins include GPCRs and Ion Channels.
MD simulations of membrane proteins allow for the characterization
and identification of interactions between the protein and the lipid
bilayer, as well as giving insight into their functions while present in
their native environment.[2] Having a membrane present in a simulation
containing a membrane-bound protein can be beneficial for reproducing
the experimental protein’s environment which can positively impact some
of the protein’s tertiary structures characteristics, such as stability.
Why run Molecular Dynamics simulations in Flare?
Flare
removes the technical barriers of command-line interfaces and allows
users to set up molecular systems using intuitive dialogs and the 3D
viewer. Users can monitor their system throughout the set-up process
using the 3D viewer and modify parameters using simple textboxes and
dropdown menus.
Flare also collates several PDB sources, allowing downloading and
preparation of the desired protein structures within the application. MD
trajectories are saved as a widely used file format and can be viewed
and analyzed with ease within the application. In this way, Flare helps
you keep all aspects of your MD experiment in one easy to navigate
place.
Setting up Molecular Dynamics simulations in Flare
Setting up and initiating MD simulations is easy and intuitive thanks
to the user-friendly GUI. Begin by downloading your PDB file from the
OPM (Orientation of Membrane Proteins) [3] server, which will give an
indication to where the upper and lower membrane leaflets will sit with
regards to the protein.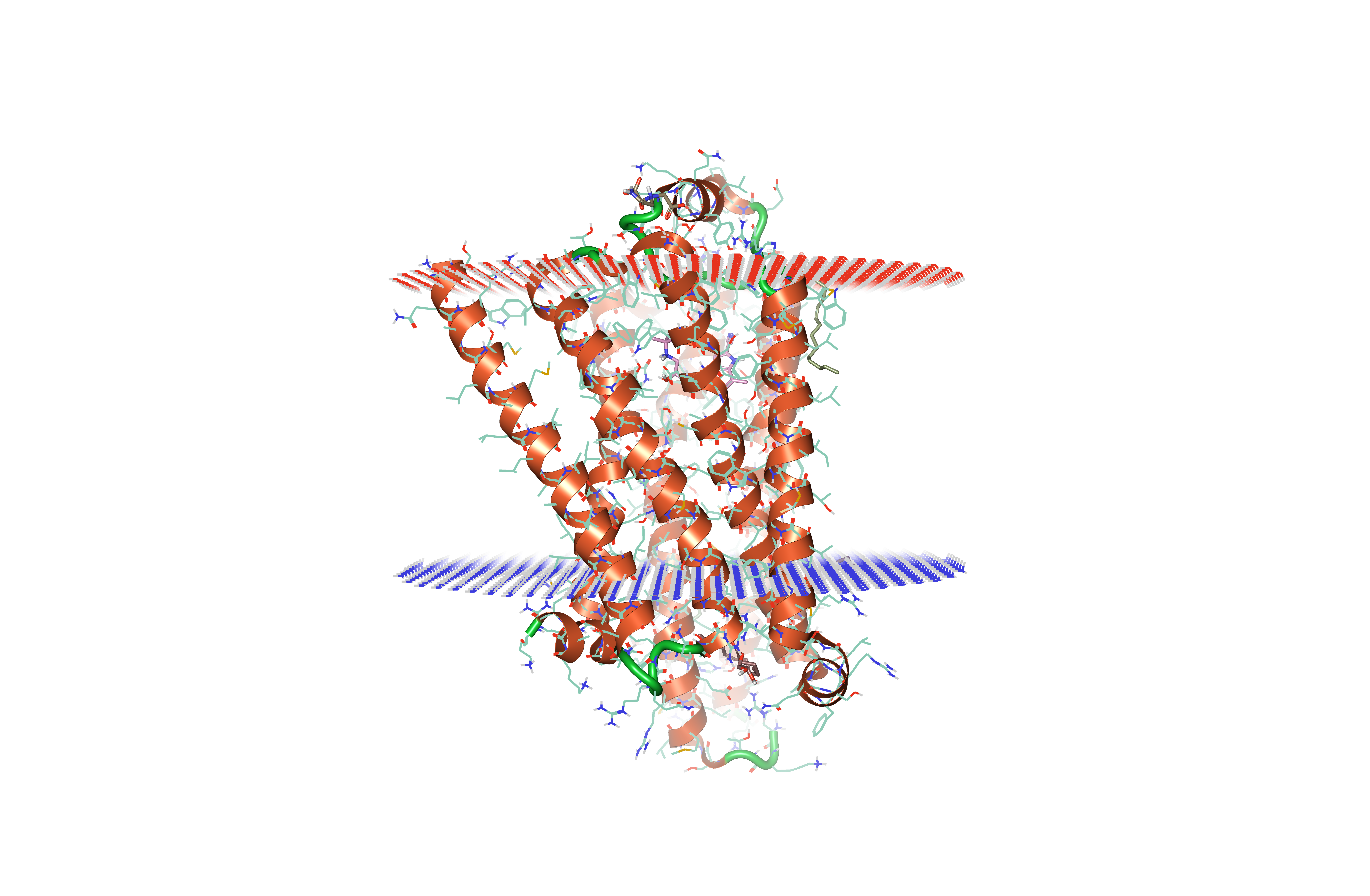
Figure 1. Avian β1-adrenoceptor (PDB: 5a8e) with OPM membrane leaflet indications.
Once your protein has been downloaded,
prepared, and undergone any manual modifications, Dynamics can be
selected from the Protein tab in the GUI.
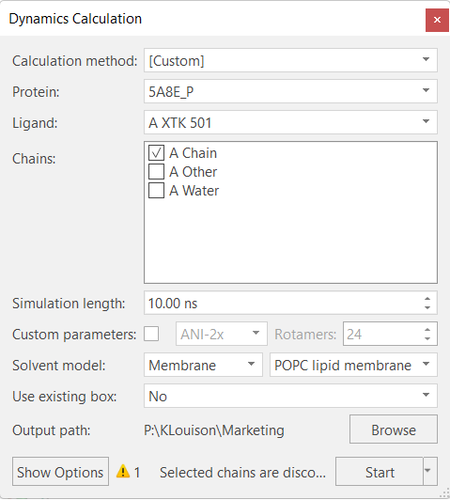
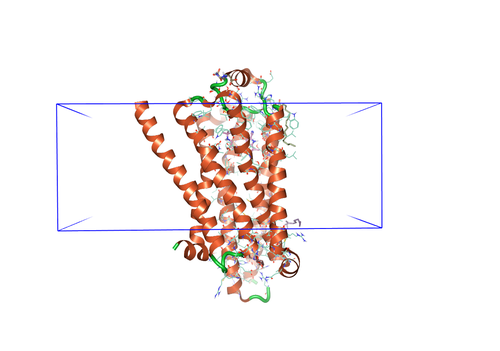
Figure 2. (Left) Example dialog box for
MD calculations in Flare. (Right) Protein with the membrane leaflet
indication in the form of a blue-outlined box.
Figure 2 (left) shows the initial dialogue box for running an MD
simulation in Flare. From this, the user is able to select which
protein, ligand, chain, and solvent model they’d like to run an MD
simulation on, as well as the simulation length in nanoseconds. Advanced
options can be accessed by clicking ‘Show Options’ at the bottom left
of the dialogue box. The advanced options allow the user to modify
parameters such as forcefields, box dimensions and time steps. The full
range of customizable parameters can be found in the Flare user manual.
After clicking ‘Start’, Flare begins ligand parameterization
(allowing for the implementation of user-defined custom parameters, if
using OpenFF and forcefield initialization. Cresset currently supports
OpenFF and AMBER forcefields. Once these steps are complete, Flare
launches the MD calculation.
Analyzing Molecular Dynamics simulations in Flare
Upon completion of the MD simulation, the trajectory .dcd file can be
analyzed from within the Flare GUI. Common analysis tools such as
time-series plots of RMSD, RMSF, Potential energy and specific
user-defined distances between atoms/residues are readily available, as
well as contacts, secondary structure and cluster analysis.
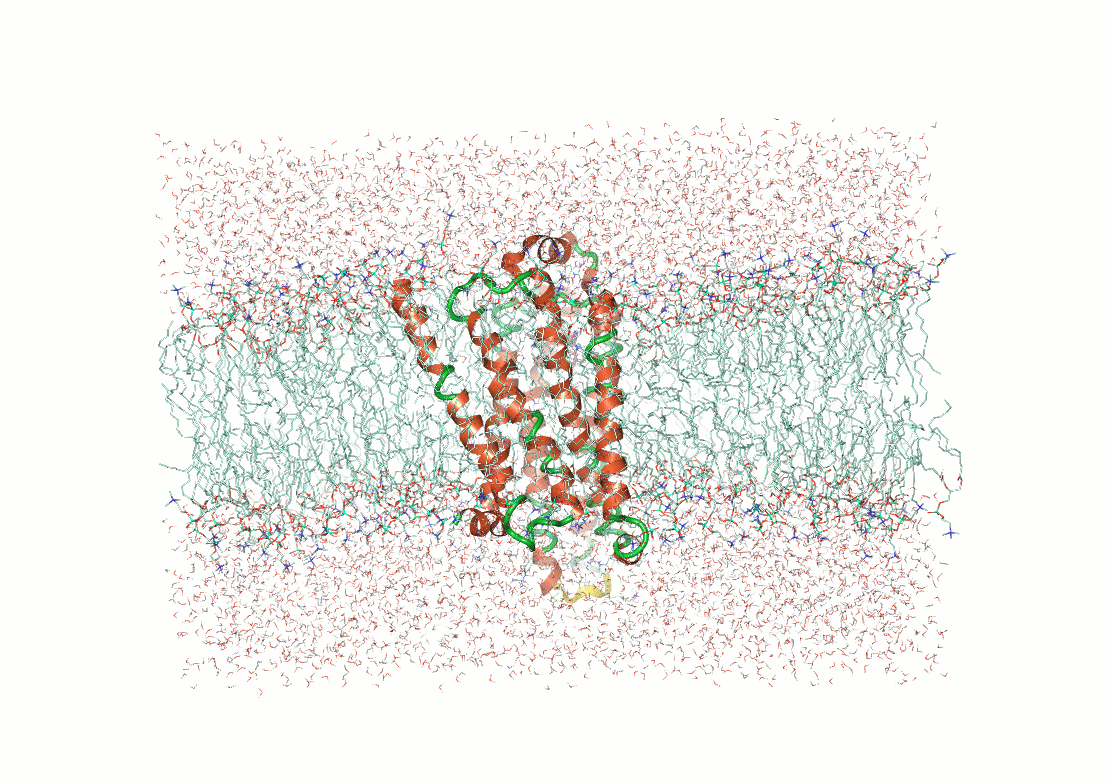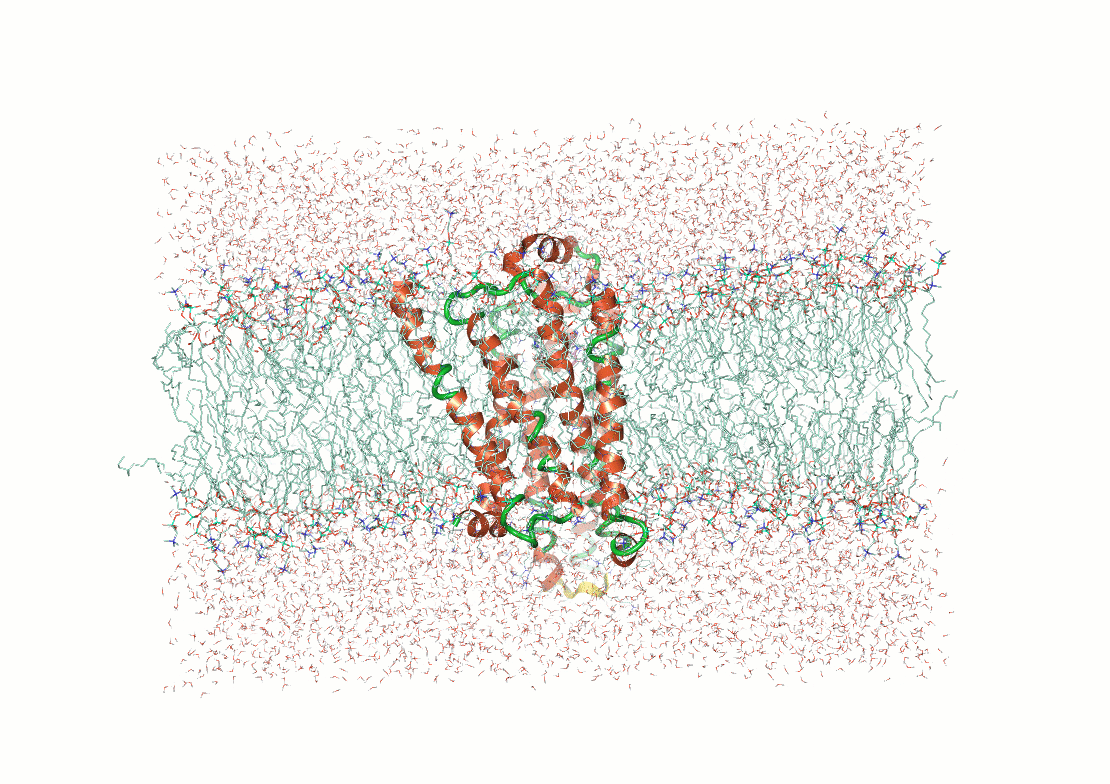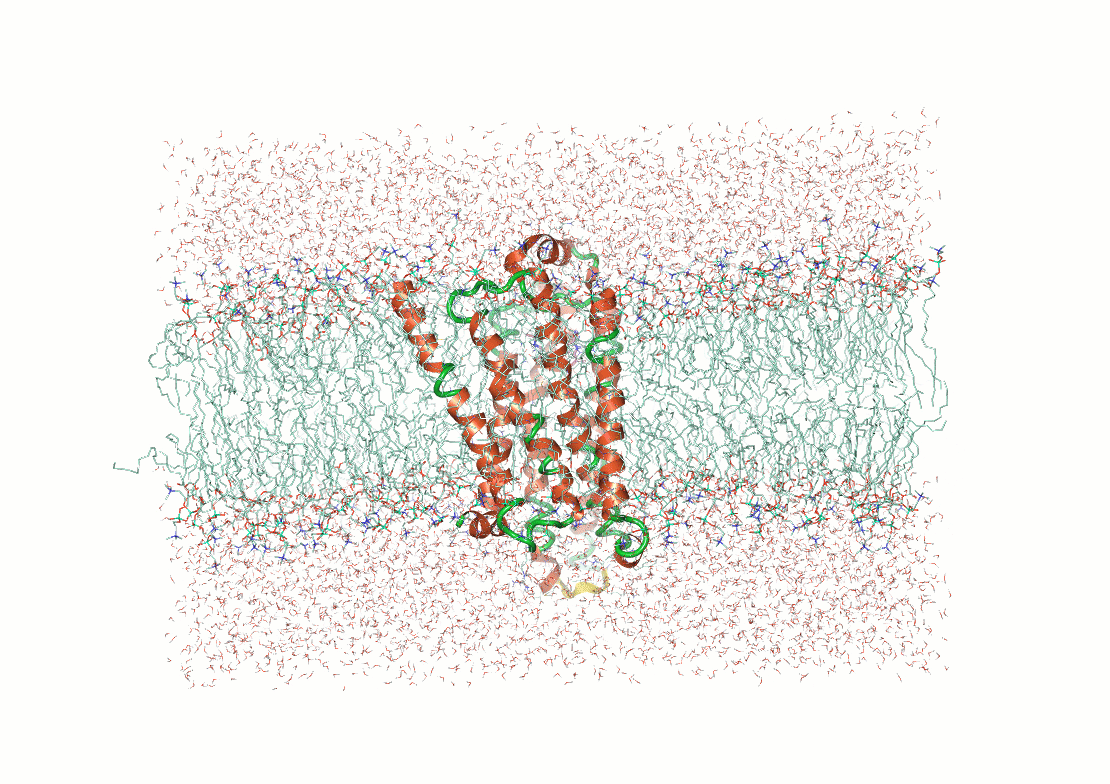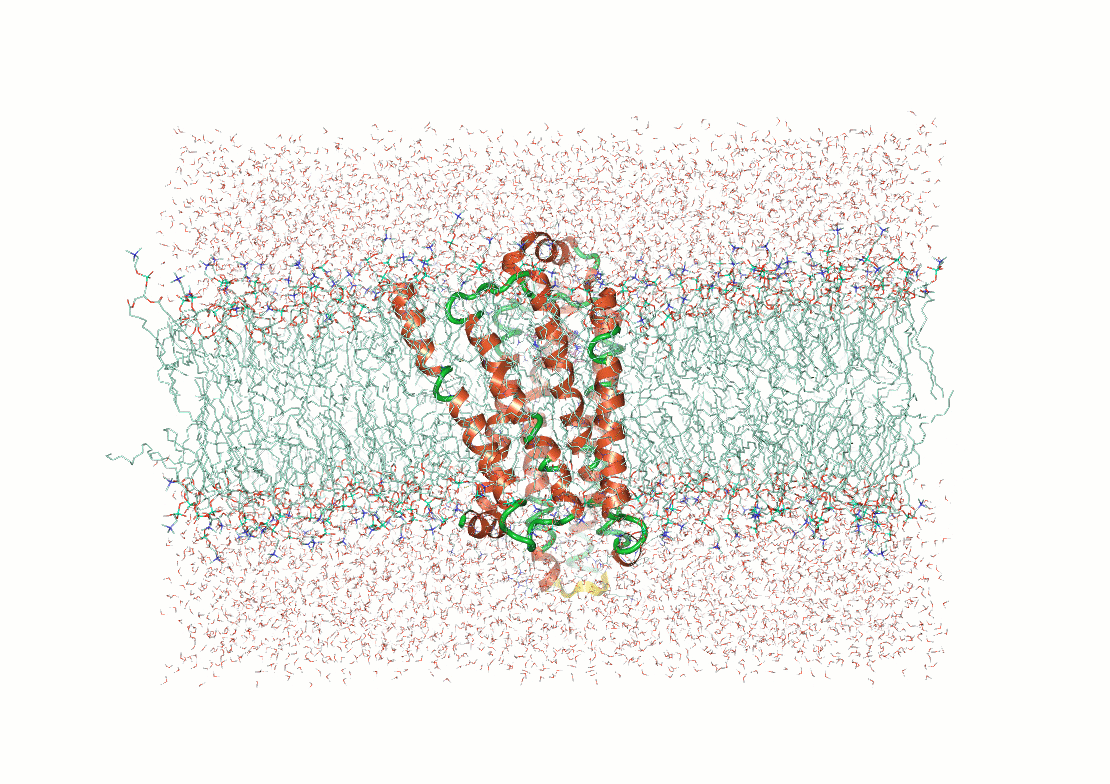
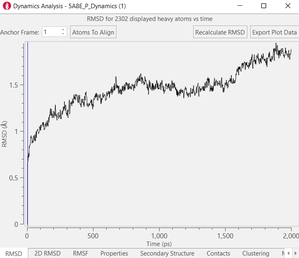
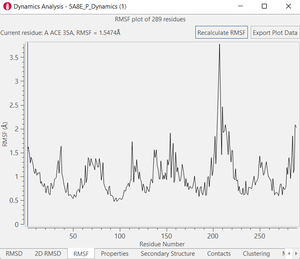
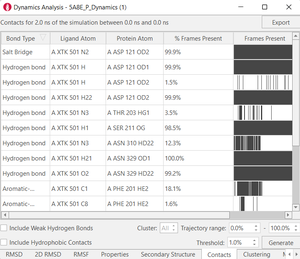
Figure 3. Examples of post calculation
analysis: (top) visual inspection (bottom left) RMSD plot, (bottom
middle) RMSF of a user-specified residue, (bottom right) protein-ligand
contacts.
Try Molecular Dynamics in Flare on your project
Run membrane protein MD in your own research project – request an evaluation of Flare.
References:
(1) Chavent, M.; Duncan, A. L.; Sansom, M. S. P. Molecular
Dynamics Simulations of Membrane Proteins and Their Interactions: From
Nanoscale to Mesoscale. Current Opinion in Structural Biology. Elsevier
Ltd October 1, 2016, pp 8–16. https://doi.org/10.1016/j.sbi.2016.06.007.
(2) Stansfeld, P. J.; Sansom, M. S. P. Molecular Simulation
Approaches to Membrane Proteins. Structure. November 9, 2011, pp
1562–1572. https://doi.org/10.1016/j.str.2011.10.002.
(3) Lomize, M. A.; Pogozheva, I. D.; Joo, H.; Mosberg, H. I.;
Lomize, A. L. OPM Database and PPM Web Server: Resources for
Positioning of Proteins in Membranes. Nucleic Acids Res 2012, 40 (D1).
https://doi.org/10.1093/nar/gkr703.