Abstract
Medicinal chemistry has renewed its interest in targeted covalent inhibitors as shown by the number of new covalent compounds on the market or in development. The discovery of the covalent inhibitor of Bruton’s Tyrosine Kinase (BTK) Evobrutinib1 will be used as the basis to demonstrate how the covalent docking workflow within Flare™2 can be used to aid design and prioritization of compounds.
Introduction
Evobrutinib is a recent BTK covalent inhibitor for the treatment of multiple sclerosis. It shows good kinase selectivity, specifically towards the EGFR and Tec family kinases, as well as a promising overall ADME profile.
Merck used the protein structure of a competitive BTK inhibitor B43 bound in active site of PDB:3GEN (Figure 1) as the starting point for the discovery of Evobrutinib. In this case study, we will also use this structure to show how the covalent docking workflow within Flare can be used to streamline design and prioritization of compounds.
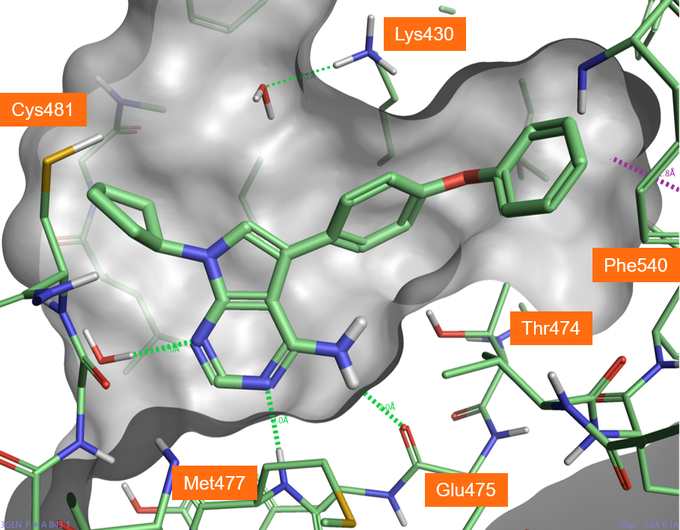
Figure 1. Crystallographic compound B43 bound in BTK active site (PDB: 3GEN), hydrogen bonds shown in green dashed lines.
Covalent docking in Flare
Covalent docking is a module in Flare using the Lead Finder™ docking algorithm3 for predicting the binding pose and interactions of covalent inhibitors. To run a covalent docking experiment in Flare, the ligand to be docked must be drawn in an appropriate reactive electrophilic form (also called a covalent warhead). These are electrophilic groups of low chemical reactivity that, once the ligand has non-covalently bound to the target protein, are positioned near a specific nucleophilic residue in the active site to which they react rapidly to form a bond. The covalent warheads supported by Flare are shown in Figure 2 – left. This list can be easily expanded with the inclusion of additional custom definitions.
A nucleophilic amino acid in the active site (either Cys, Lys, Ser or Tyr) must also be specified to start the experiment.
The covalent docking workflow then performs a chemical transformation to the input compound which modifies the covalent warhead to generate a pseudo-covalent ligand. This is not formally covalently bonded to the protein but is bonded to the nucleophilic atom from the selected amino acid which is protonated (Figure 2 – right). 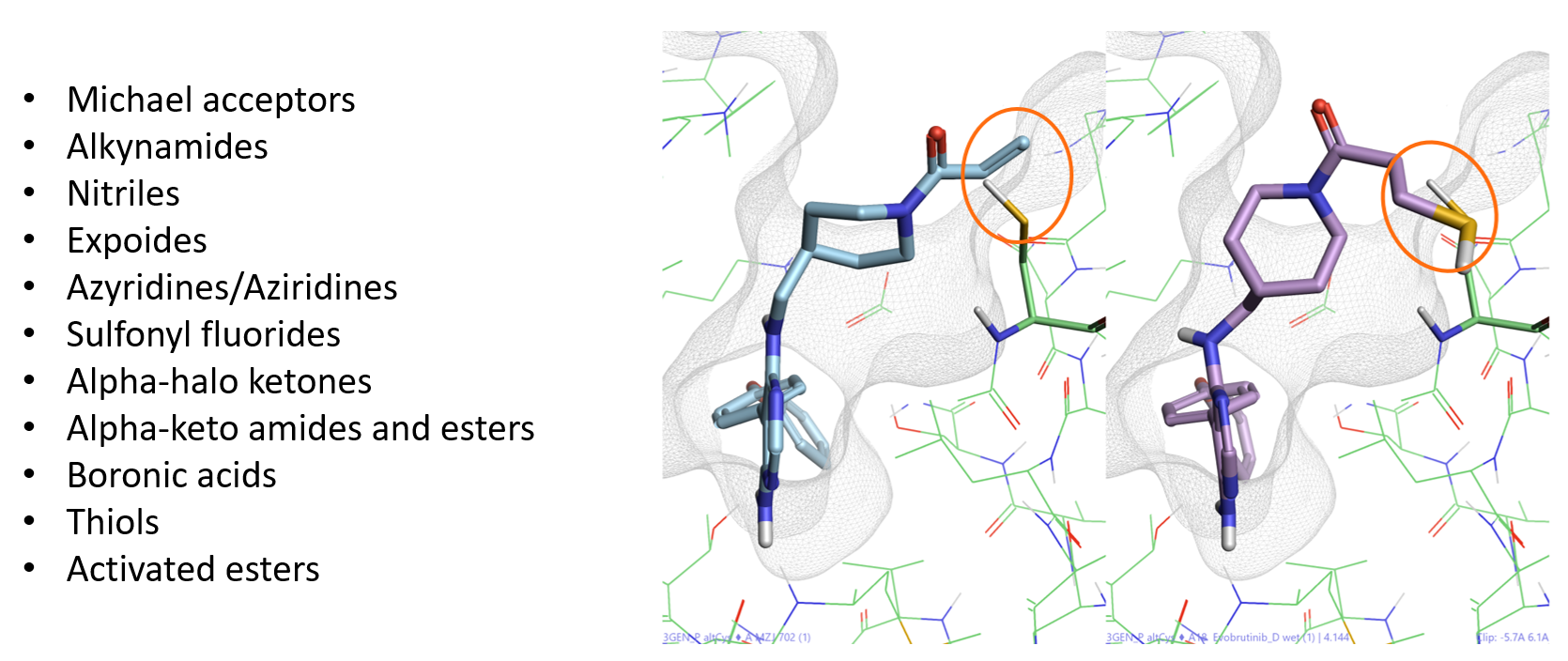
Figure 2. Left: List of covalent warheads supported by Flare. Right: Chemical transformation for Evobutrinib covalently docked to Cys481.
Preparation of protein, ligands and docking
The PDB:3GEN protein-ligand crystal structure was downloaded in Flare and fully prepared to add hydrogen atoms, optimize hydrogen bonds, remove atomic clashes, and assign optimal protonation states. The prepared system was further refined by manually rotating the hydroxyl of Threonine 474, to create a more favorable ligand-protein H-bond interaction, also in line with other BTK protein structures.
As the PDB:3GEN protein structure (the starting point for the discovery of Evobrutinib in Caldwell et al.1) is not bound to a covalent ligand, the side chain of the target nucleophilic amino acid Cys481 may not be in a suitable conformation to form a covalent bond. To explore this potential issue, a duplicate protein was created and modified by choosing an alternative rotamer for Cys481: the modified amino acid was then minimized. Both proteins were used to explore the effect of choosing alternative rotamers for the target nucleophilic residue on docking.
Compounds A3 to A26 from Caldwell et al.1 were imported into Flare applying Flare’s rule-based protonation (Figure 3). The protonation state of the imported ligand structures was carefully examined, and the pyridyl aza nitrogen of compound A5 (an amino-pyridine hinge binder) was manually neutralized to ensure consistent binding poses and maintain its ability to form a H-bond with the backbone Met477 NH. All the ligands in this dataset (including A18, Evobrutinib) carry a Michael acceptor as the covalent warhead.
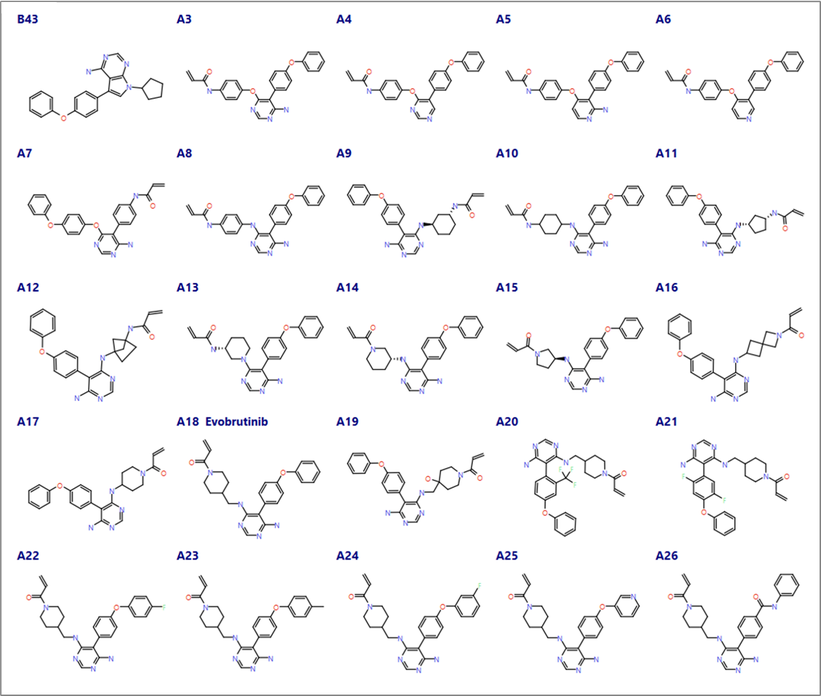
Figure 3. Compounds from Caldwell et al.1 with different cores exploring hinge binding domain (A3-A8), alternative linkers between the warhead and the core (A9-A19) and the selectivity pocket (A20 -A26).
Preliminary docking experiments
Exploratory docking of the hinge binding designs (A3-A8) within the active site of 3GEN was performed using the ‘Normal’ covalent docking method in Flare, excluding the crystallographic waters and defining Cys481 as the nucleophilic amino acid residue, as shown in Figure 4. This initial experiment was performed to quickly verify whether additional constraints where needed, or changes required to the ligands or protein, to obtain good docking results.
The poses from each docking experiment were prioritized using the Lead Finder ‘rank score’, based on a semi-empiric molecular mechanical functional and optimized to identify the crystallographic poses.
As the results were promising, further preliminary covalent docking experiments were ran using the ‘Accurate but Slow’ and ‘Very Accurate but Slow’ methods to optimize conformational sampling and maximize the consistency of results for some of the other designs containing more flexible linkers. The ‘Accurate but Slow’ method was the best balance between speed and accuracy.
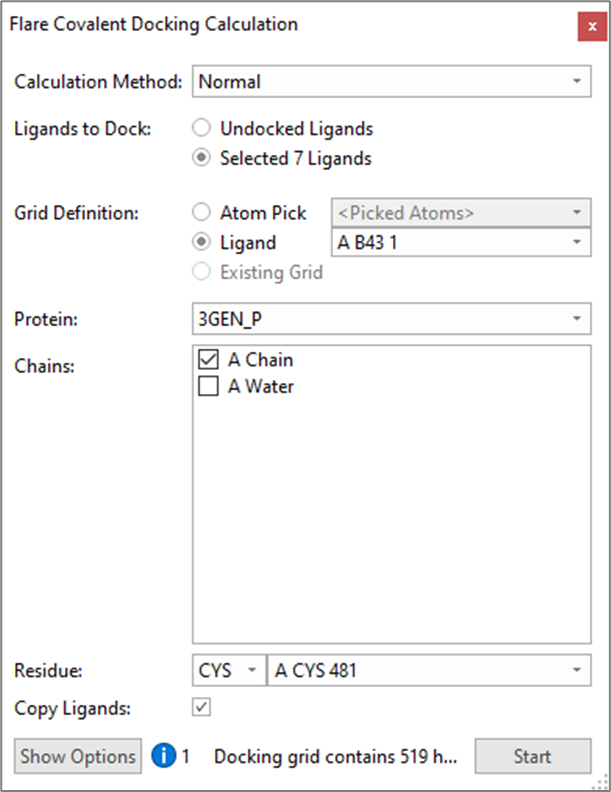
Figure 4. Flare Covalent Docking Calculation panel.
Results and analysis
For the hinge binder designs (A3-A8 in Figure 3) the best results came from using the original Cys481 rotamer, these designs are close analogues of the crystallographic ligand B43. The covalent docking workflow in Flare scored compounds A3 and A5 the best and A7 the worst.
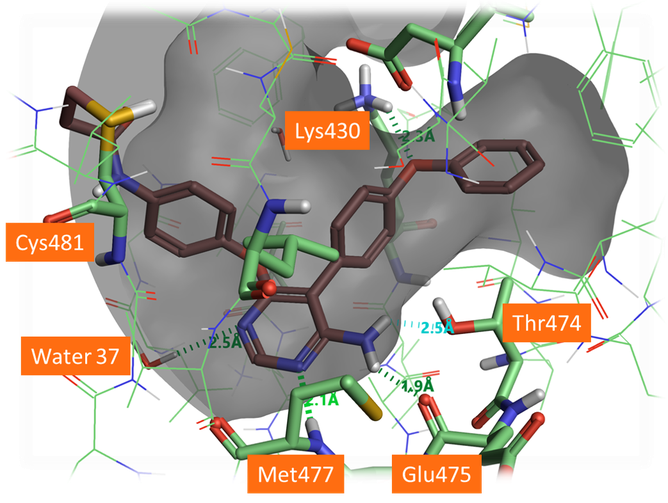
Figure 5. Docked pose of A3 forming H-bonds with the protein backbone and the gate keeper Thr474. The aza-nitrogen in the 5 position shows a H-bond to a crystallographic water in the active site which was not included in this docking. It would be beneficial for this water to be included in docking for this chemical series.
As shown in Figure 5, A3 and A5 form good H-bonding to the backbone amine of Met477, the backbone carbonyl of Glu475 and the hydroxyl side chain of Thr474, the so-called gate keeper residue. As the gate keeper residue in EGFR kinases is instead methionine 790, forming H-bond with the Thr474 residue offers the opportunity to gain in intrinsic selectivity for BTK over EGFR kinases. The phenoxyl phenyl moiety fills the selectivity pocket making a H-bond with Lys430. Interestingly the 5-aza nitrogen of the pyrimidine ligands does not interact directly with the protein but makes a H-bond interaction with a crystallographic water as the original B43 compound. Including this water molecule in future docking experiments for the pyrimidyl hinge binding series would be advantageous to identify and score the most promising poses.
Compounds A4 and A6 lack the 2-amino substituent to form H-bonds with Thr474, but the other interactions are the same as those observed for compounds A3 and A5. However, as the aim of this drug discovery effort was to target good kinase selectivity, not forming interactions with Thr474 in BTK could impact the intrinsic selectivity for BTK vs EGFR.
A7 carries a reversed amino-pyrimidine moiety which is unable to form the same interactions as the other designs in this set with the hinge domain, in line with its lower activity for BTK and EGFR reported by Caldwell et al.1.
Covalent docking of the linker designs (A9-A19 in Figure 3) generated good poses for both the original and modeled Cys481 rotamers. Most of the compounds show a similar pose across all docking experiments with differences observed only in the linker conformation. These designs are harder to prioritize as this region of the active site is open and predominantly solvent accessible, as such the insights from docking were limited with compounds A11 and A13-15 scoring poorly when using the original Cys481 rotamer and A13 and A16 scoring poorly for the alternative Cys481 rotamer. Duplicate poses were minimized in the absence of the protein to help infer strain within the compounds.
The designs for the selectivity pocket also focused on improving the microsomal stability of the phenoxy phenyl rings. The docking scores and the protein ligand contacts identified some steric clashes from substituents on the phenyl rings of A20 and A21 using either Cys481 rotamer. The other compounds in this set A22 to A26 have changes to the terminal phenyl ring and all fit within the selectivity pocket.
As with conventional docking having the protein prepared and in the correct conformation has significant an impact on the results. In this case study the ligands that where less flexible tended to prefer the original Cys481 rotamer such as the hinge binder designs which contained aromatic linkers between the core and the warhead. More flexible ligands in the linker designs generated better docking poses when using the alternative Cys481 rotamer.
Comparison of docked Evobrutinib with the X-ray structure
A comparison of the crystal structure of Evobrutinib covalently bound to the BTK active site (PDB: 6OMU) with the docked pose of Evobrutinib within the original PDB: 3GEN active site highlights a small number of differences between the two crystal structures (Figure 6). In particular, the conformation of the side chain of Cys481 in 6OMU is indeed different to that of 3GEN: there are also some minor differences in the conformation of Lys430 and some additional crystallographic waters in the active site of 6OMU.
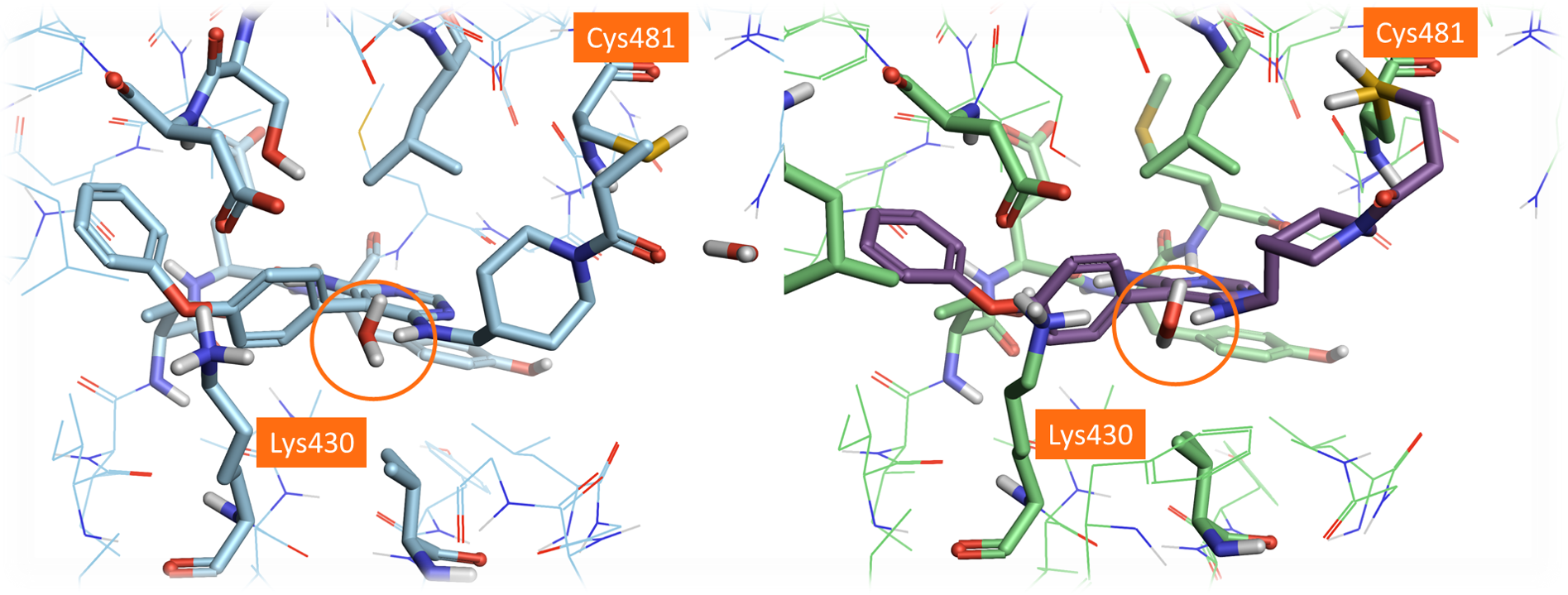
Figure 6. Left: Crystal structure of Evobrutinib (PDB:6OMU). Right: the best scoring pose of Evobrutinib docked in the dry active site of PDB: 3GEN using the ‘Accurate but Slow’ method. The different side chain positions of Cys481 can be observed by comparing these two structures.
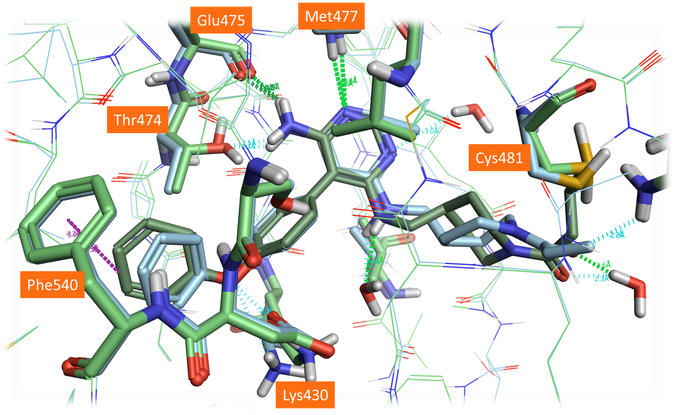
Figure 7. Crystal structure (PDB: 6OMU, light blue) and the best scoring pose of Evobrutinib (green) using an alternative Cys481 rotamer position and including waters in the active site.
The alternative modeled side chain conformation for Cys481 is closer to its experimental position in 6OMU (Figure 7). Not surprisingly, choosing a different rotamer for the nucleophilic residue affects the entire binding pose of the compound: using the alternative rotamer for Cys481 improves the results significantly, but the linker conformation is still slightly different to that observed in the crystal structure. This could be further improved by including selected water molecules in the active site from the 3GEN protein structure, specifically the ‘1024’ water molecule near the amine in the linker chain (Figure 7). This final pose is very close to the observed crystal structure, with only minor differences between the saturated ring conformation.
Conclusion
Flare provides a user friendly interface to prepare, design and dock your covalent ligands. The docking workflow generates pseudo-covalent ligands which can be saved in the Flare project as conventional docking results, facilitating the exploration of the different docked poses and the further post-processing of the results to select the best candidates for further progression in the drug discovery project.
References and links
- Caldwell et al., J. Med. Chem. 2019, 62, 7643-7655
- https://www.cresset-group.com/software/flare/
- https://www.cresset-group.com/software/lead-finder/